Test Name
Legionella pneumophila PCR (LEGPCR)
CPT Codes
87541
Methodology
Polymerase Chain Reaction (PCR)
Turnaround Time
3 days
Specimen Requirements
Specimen Type:
Bronchoalveolar lavage (BAL)
Volume:
3 mL
Minimum Volume:
2 mL
Collection Container:
Sterile specimen container
Transport Temperature:
Ambient
Alternative Specimen
Specimen Type:
Sputum, induced
Aspirate, tracheal
Volume:
1 mL
Collection Container:
Sterile specimen container
Transport Temperature:
Ambient
Stability
Ambient:
24 hours
Refrigerated:
7 days
Frozen:
30 days
Reference Range
No Legionella pneumophila detected
Additional Information
Background Information
Legionella pneumophila is the most common pathogenic species of the 42-recognized Legionella species and is associated with significant mortality in elderly patients and those with severe underlying disease. Diagnostic delay also may result in increased mortality. Target sequences within the genes that encode the 5S and 16S ribosomal subunits, as well as the MIP (macrophage infectivity potentiator) gene, in conjunction with real-time PCR, are useful for the detection of the Legionella genus and, specifically, the species L. pneumophila.
The MIP gene, which encodes a 24-kDa protein virulence factor that facilitates the entry of legionellae into amoebae and macrophages, has sufficient sequence variability between the Legionella species to allow for the specific detection of L. pneumophila by real-time PCR. This assay has 100% specificity and sensitivity.
Clinical Information
Multiple laboratory methods should be employed to ensure the diagnosis of Legionnaire’s disease (LD), a bacterial pneumonia caused by L. pneumophila (90% of cases) or other Legionella species. Molecular methods are more sensitive than culture for the diagnosis of LD.
The PCR assay performed at Cleveland Clinic will not detect disease caused by Legionella species other than L. pneumophila.
Culture for Legionella species from respiratory sites is a sensitive (~80-90%) method for diagnosing severe, untreated disease, but insensitive (~20%) for the diagnosis of mild disease. Specimens from non-respiratory sites should not be submitted for Legionella culture unless there is a high index of clinical suspicion to support the request. Urine antigen assays for L. pneumophila serogroup 1 will be positive in ~90-95% of patients with severe disease due to the Pontiac monoclonal subtype of serogroup 1, but positive in only 50% of these patients with mild disease. The urine antigen assay is unreliable for the diagnosis of severe LD caused by L. pneumophila other than serogroup 1 or a different Legionella species (detects less than 5-40% of cases).
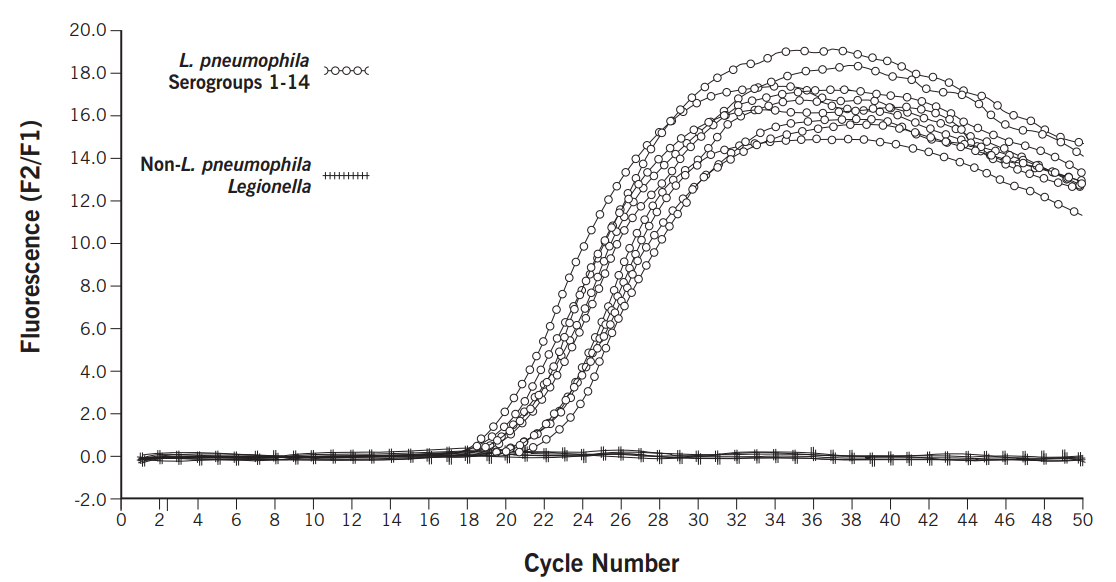
Interpretation
PCR results are reported qualitatively as positive or negative for Legionella pneumophila.
Limitations
This assay does not contain an internal amplification control; instead, the false-negative rate, which to date has been 0%, is monitored according to the College of American Pathologists’ guidelines.
This assay only detects L. pneumophila, as Legionella species other than L. pneumophila infrequently cause legionellosis. The implementation of this test was intended to replace the less-sensitive direct immunofluorescence assay, and it is intended to be used in conjunction with culture.
Methodology
The LightCycler FastStart DNA Master Hybridization Probe Kit (Roche Diagnostics, Indianapolis, Ind.) is used in conjunction with species-specific Legionella pneumophila primers and probes. The PCR is performed on the LightCycler system (Roche).
Qualitative positive and negative results are determined based on analysis of the amplification curves and post-amplification melt curve.
Suggested Reading
1. Wilson DA, Yen-Lieberman B, Reischl U, Gordon SM, and Procop GW. Detection of Legionella pneumophila by Real-Time PCR of the MIP Gene. J Clin Microbiol. 2003;41:3327-3330.

