Test Name
Hepatitis C Genotype (HEPGEN)
CPT Codes
87902
Methodology
Reverse Transcription/Polymerase Chain Reaction (RT/PCR)
Turnaround Time
5 – 7 days
Specimen Requirements
Type:
Plasma
Volume:
3 mL
Minimum Volume:
1 mL
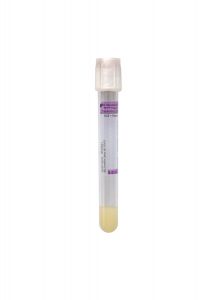
Tube/Container:
White BD Hemogard™ K2EDTA Plasma Preparation Tube
Transport Temperature:
Refrigerated
Alternative Specimen
Type:
Plasma
Volume:
3 mL
Minimum Volume:
1 mL
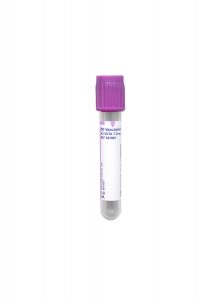
Tube/Container:
Lavender BD Hemogard™ K2EDTA Tube
Transport Temperature:
Refrigerated
Alternative Specimen
Type:
Serum
Volume:
3 mL
Minimum Volume:
1 mL
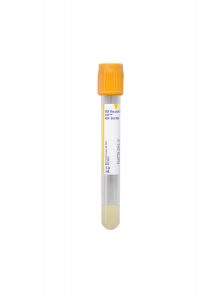
Tube/Container:
Gold BD Hemogard™ Serum Separation Tubes (SST)™
Transport Temperature:
Refrigerated
Stability:
Plasma
Ambient:
3 days
Refrigerated:
3 days
Frozen:
60 days
Stability:
Whole blood
Ambient:
15-30°C for up to 6 hours prior to centrifugation
Refrigerated:
2-8°C for up to 6 hours prior to centrifugation
Frozen:
Unacceptable
Reference Ranges
Capable of discriminating among HCV subtypes 1a, 1b, 2a-2c, 3a-3c, 4a-4h, 5a, and 6a
Additional Information
Background Information
Chronic infection with the hepatitis C virus (HCV) remains one of the world’s most important clinical and public health problems. It has been estimated that approximately 3% of the world’s population is infected with HCV, which represents nearly 170 million people worldwide.1,4 In the United States, up to 3.9 million people (1.8% of the population) are currently living with HCV, of which as many as 2.7 million suffer from chronic infection.2,3 In the Western world, chronic damage from hepatitis C is the primary cause for end-stage liver disease that requires liver transplantation.
Clinical Significance
The 1989 discovery of the hepatitis C virus was a major development. Previously, it was clear that a major cause of acute hepatitis after a blood transfusion was related to neither hepatitis A nor to hepatitis B. This resulted in the early name for this disease: non-A, non-B hepatitis. It is now known that HCV is the cause for most of the non-A, non-B hepatitis cases.
HCV is a single-stranded RNA virus that has approximately 9400 nucleotides that demonstrate significant genetic variation. HCV replicates in the liver and is detectable in serum during acute and chronic infection. The RNA polymerase lacks the proofreading functions of DNA polymerase and introduces random nucleotide errors. This results in a relatively high rate of spontaneous nucleotide substitutions. As a consequence, HCV is a highly-heterogeneous virus with at least six known major genotypes and more than 80 subtypes identified worldwide.5 Genotypes are classified based on differences in the amino acid sequence of specific proteins.
Recent understanding of the natural history of chronic hepatitis C has greatly expanded, and more effective therapeutic strategies have been developed. Several studies have shown a strong correlation between HCV genotype and a patient’s response to various treatments. Determining genotypes is necessary because some hepatitis C viruses with certain genetic variations are harder to treat successfully and usually require a different treatment approach; others are much easier to treat and respond well to shorter treatment schedules.
For many years, the standard of care for patients with chronic hepatitis C infection was peginterferon (PegIFN) alfa and ribavirin (RBV) taken for 24 to 48 weeks, depending on the genotype; however, less than 50% of patients responded to this therapy. In May 2011, the FDA approved the use of two new protease inhibitors – Incevik (telaprevir) and Victrelis (boceprevir) – for the treatment of hepatitis genotype 1, the most common form of hepatitis in North America. The drugs, used in conjunction with the standard interferon and ribavirin therapies, represent the first major therapeutic advance in the treatment of hepatitis C in more than a decade. The new protease inhibitors block the replication of an enzyme that is crucial for the hepatitis C virus to reproduce. In patients with hepatitis genotype 1, the use of the new protease inhibitor combined with the traditional antiviral treatment eliminated the virus in 70 to 80% of all cases. Patients with the other types of hepatitis C continue to be treated with peginterferon and ribavirin, which is successful in 80% or more in those with genotypes 2 and 3 infections.6-8
Clinical Indications
Pre-treatment analysis of hepatitis C genotype is used to determine the duration of therapy and predict therapeutic response.
Interpretation
The predominant HCV genotypes in the United States are 1a, 1b, 2a, 2b, and 3a; the other subtypes (4, 5, 6) are more prominent in other parts of the world.9
Limitations
This assay is based on the sequence variability of the 5’UTR, allowing differentiation between HCV genotypes 1 to 6. Subtyping may occasionally be limited.
Methodology
The gold standard for genotyping is determining the nucleotide sequence of an HCV isolate. Currently, this method is not practical for the clinical diagnostic laboratory, and a less labor-intensive technique, such as Line Probe Assay (LiPA), is employed.
The LiPA method uses genotypic-specific oligonucleotide probes immobilized onto membrane strips. The end products obtained from RT-PCR of the 5’UTR region of the clinical isolate are then hybridized onto the membrane containing the immobilized-oligonucleotides. A purple-brown line develops where sequence homology occurs between the biotinylated PCR products and the probe. Hybridization of 5’UTR amplification products with genotype-specific probes is capable of discriminating among HCV subtypes 1a, 1b, 2a-2c, 3a-3c, 4a-4h, 5a, and 6a.10
References
1. Organization WH. Hepatitis C surveillance and control. Available at: http://www.who.int/=csr/disease/hepatitis/whocdscsrlyo2003/en/index4.html#incidence.
2. Armstrong GL, Wasley A, Simard EP, McQuillan GM, Kuhnert WL, Alter MJ. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Intern Med. 2006;144:705-14.
3. Centers for Disease Control. www.cdc.gov/hepatitis/PDFs/disease_burden.pdf.
4. WHO Hepatitis C fact sheet. 2011.
5. Davis GL. Hepatitis C Virus genotypes and quasispecies. Am J Med. 1999;107(6B):21S-25S.
6. Poordad, F, et al. (March 2011). “Boceprevir for Untreated Chronic HCV Genotype 1 Infection”. N Engl J Med. 364(13):1195 206. doi:10.1056/NEJMoa1010494. PMID 21449783.
7. Bacon, B, et al. (March 2011). “Boceprevir for Previously Treated Chronic HCV Genotype 1 Infection”. N Engl J Med. 364(13):1207-17. doi:10.1056/NEJMoa1009482. PMC 3153125. PMID 21449784.
8. McHutchison JG, Manns MP, Muir AJ, et al. (April 2010). “Telaprevir for previously treated chronic HCV infection”. N Engl J Med. 362 (14): 1292–303. doi:10.1056/NEJM oa0908014. PMID 20375406.
9. Mahaney K, Tdeschi, V, Maertens G, et al. Genotypic analysis of hepatitis C virus in American patients. Hepatology; 1994; 20:1405-1411.
10. Stuyver L, Wyseur A, Van Amhem W et al. Second generation line probe assay for hepatitis C virus genotyping. J Clin Microbiol. 1996; 24:2259-2266.